Sub Programme Area 301
Salinity tolerance is a complex and multigenic trait in plant species. Rice, among cereals, is most sensitive to salinity. In this subtheme, mechanisms of tolerance in the naturally halophytic wild rice species Oryza coarctata are examined. In addition, natural variation in salinity tolerance seen in coastal cultivated rice landraces (Oryza sativa L.) is analyzed.
301.1 Studies Related to Salinity Tolerance in Oryza coarctata
Analysis of Salt-Secretory Microhairs of O. coarctata: Halophytic, perennial wild rice O. coarctata leaves show the presence of microhairs on their leaf surface furrows that secrete salt under salinity. An in vitro secretory system for O. coarctata leaf tissue under salinity established previously was used to examine the effect of plant hormones on leaf secretory volume and Na+ content in secretions from the adaxial leaf surface. An efficient method for the isolation and enrichment of O. coarctata microhairs has been reported previously. The robustness of the microhair isolation procedure was confirmed by generating a transcriptome from microhairs under salinity conditions. A total of 551484 unique transcript sequences with average length of 631 bp were obtained. Functional annotation revealed the presence of transcripts involved in diverse cellular processes, including 310 transcription factor (TF) related transcripts and 1075 transporter-related genes in O. coarctata microhairs. Moreover, 3530 differentially regulated transcripts under salinity were identified in the study.
Transcriptomics (RNAseq) of O. coarctata Leaf and Root Tissues: Oryza coarctata leaf and root tissue specific transcriptomes (RNAseq) generated previously were analyzed in depth. A total of 39878 sequences were annotated and 109716 gene ontology terms assigned. Of these, 7117 sequences were mapped onto the database of the Kyoto Encyclopedia of Genes and Genomes (KEGG). In leaf tissues under salinity, 206 upregulated transcripts were identified; these included several novel, uncharacterized transcripts, in addition to an ethylene responsive factor (ERF)-like TF gene. Also, 63 transcripts were downregulated, including those coding for auxin-responsive protein, nitrate reductase and benzothiadiazole (BTH)-induced ERF-transcriptional factor 4. In salinity-treated root tissues, 153 transcripts were upregulated, including several coding for uncharacterized proteins, a dehydration responsive protein and a DEAD-box ATP-dependent RNA helicase. Moreover, 880 transcripts in roots showed downregulation of expression, including those coding for 3-ketoacyl-CoA synthase, hsp90 and GHR1 protein. RT-qPCR validation of these up- or downregulated transcripts were completed and found to correlate with RNAseq data.
Examining Apoplastic Barriers in O. coarctata Roots under Salinity: Oryza coarctata rice species have an elaborate rhizomatous system from which arise thin rhizoid-like roots that help in absorption and anchorage. Microscopic examination of soil-grown O. coarctata roots has previously shown strong apoplastic lignified/suberized exodermal and endodermal layers suggestive of a role in controlling ion movement. On the other hand, freshly emerged O. coarctata roots in a hydroponic system lack these apoplastic barriers in both elongation and maturation zones and also show root hair development in the absence of salt in the medium. However, inclusion of Na+ in the medium results in the establishment of strong endodermal apoplastic barriers in the maturation zone and to a lesser extent in the elongation zone. Moreover, exodermal apoplastic barriers are established to more or less equal extent in both zones. This optimized system will now be used to examine Na+ distribution in both root zones under salinity.
Examining Lignification and Suberization in the Aerial Shoot Proximal Nodes and Internodes of O. coarctata Rhizomes under Salinity: In O. coarctata, underground rhizomatous tissues form a substantial biomass below ground that anchors the species to the shallow soil in intertidal mangrove communities. Below-ground rhizomes are an important interface in the transition to aerial shoots in O. coarctata that should, in theory, control ion movement to aerial shoots. With this background, anatomical regions of the aerial shoot and below-ground rhizome regions have been examined. Distinct anatomical changes observed in leaf proximal internodes of O. coarctata under salinity have been documented, and these correlate with reduction of apoplastic barriers under salinity and patterns of Na+ accumulation observed using the fluorescent dye CoroNa Green.
Analysis of Transporter Genes and Their Regulation in Wild Rice O. coarctata: HKT1;5 genes are important determinants of crop salinity tolerance. They encode plasmalemma-localized Na+ transporters, retrieve xylem Na+ into xylem parenchyma cells and reduce shoot Na+ accumulation. Transport characteristics of the roots and shoots expressing O. coarctata HKT1;5 (OcHKT1;5) were examined in contrast to that from cultivated rice O. sativa (OsHKT1;5) in both yeast and Xenopus oocytes. OcHKT1;5 is a low-affinity sodium transporter compared with OsHKT1;5. Using homology modelling, critical amino acid residues controlling these distinctive characteristics were identified in OcHKT1;5. Site-directed mutagenesis of OcHKT1;5 followed by transport assays of the mutagenized construct in Xenopus confirmed the importance of one key amino acid change in conferring this low-affinity sodium-transporting property in OcHKT1;5. Vacuolar-localized OsNHX1 is extensively characterized in O. sativa and controls vacuolar accumulation of sodium. The genus Oryza consists of 11 diploid or tetraploid genome types (AA, BB, CC, EE, FF, GG, BBCC, CCDD, HHJJ, HHKK and KKL) with cultivated rice, diploid O. sativa, having an AA-type genome. Oryza NHX1 orthologous regions (gene organization, 5′ upstream cis elements, amino acid residues/motifs) from closely related Oryza AA genomes were found to group distinctly as compared with NHX1 regions from more ancestral Oryza BB, FF and KKLL (genome-type) species. Further, these sequence-specific differences were found to affect alternative splicing of transcripts generated from the NHX1 gene that involve two separate intron retention events occurring in the 5′ and 3′ UTR, respectively. Data shows that the IR event involving the 5′ UTR is present only in more recently evolved Oryza AA genomes while the IR event governing retention of the 13th intron of Oryza NHX1 (terminal intron) is more ancient in origin, also occurring in O. coarctata (KKLL).
301.2 Genetic Diversity of Rice Landraces from Saline Coastal Regions of India
We had previously reported about genotyping rice landraces (Oryza sativa L.) from saline coastal regions of India. The relative salinity tolerance of 43 rice landraces at seedling stage, using 13 unbiased morpho-physiological and biochemical parameters related to salinity tolerance, was assessed. Of the 43 varieties, 25 were tolerant, 15 were moderately tolerant, 1 was moderately susceptible and 2 sensitive checks were found to be highly susceptible based on the aforementioned parameters and assignment of standard evaluation scores (SESs) A high correlation was observed with Na+/K+ ratio, and cluster analysis classified the rice genotypes into six groups based on their responses to salinity. Saline-sensitive check IR29 was grouped with moderately tolerant rice landraces in Group I while more tolerant landraces were clustered in Groups III and IV. Group II had landraces that were more tolerant than those in Group I while Group VI had landraces that were more tolerant than those in Group II.
301.3 Phenotypic and Molecular Characterization of Saline-Tolerant Kagga Paddy:
The applications to recognize Kagga paddy as a saline-tolerant variety were considered favourably by the Protection of Plant Varieties & Farmers’ Rights Authority (PPV&FRA), government of India. The authority has requested additional data on the biochemical parameters for finalizing approval of the applications. Currently, the biochemical data are being generated for submission to PPV&FRA.
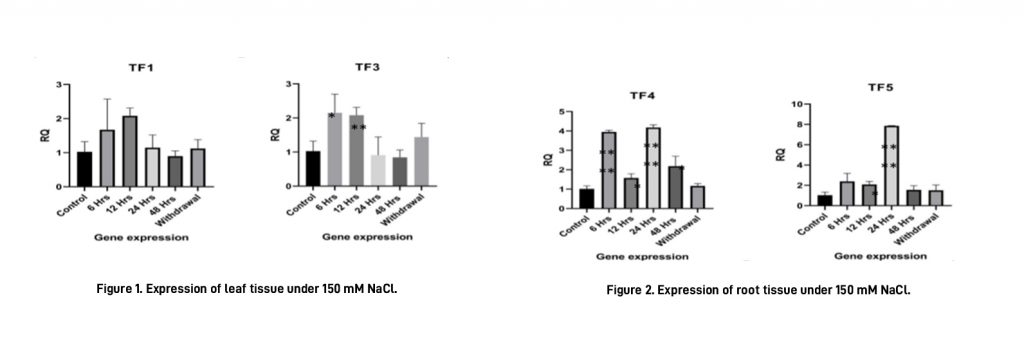
301.4 Functional Analysis of Five Drought-/Salt-Responsive Transcription Factors from the Tree Species Prosopis juliflora
The functional analysis of the abiotic responsive transcription factors for Prosopis juliflora, an extremely drought-tolerant tree species from the family Fabaceae, was continued. In the current year, the results of the expression profiling of the selected transcription factors under drought and salt stress, analysed through quantitative real-time PCR (q-RT PCR), were in alignment with the results of transcriptome profiling of P. juliflora under the same conditions in both the leaf and root tissues. All transcription factors analysed were found to be induced by drought and salt stress at at least one-time point of treatment. Withdrawal of the stress resulted in downregulation of expression of all transcription factors, except for TF5 in leaf tissue under drought stress, TF1 and TF3 in leaf tissue under salt stress, and TF1 and TF3 in root tissue under salt stress. The results of the RT PCR analysis confirm the importance of selected transcription factors in drought and salt stress tolerance (Figures 1 and 2).